Introduction
Validation is an essential step for a laboratory to implement a clinical test. Validation aims at ensuring that a test meets the diagnostic requirements. For a genetic test, validation has to address both the clinical validity of the test and the analytical validity of the measurement. Validation is an important parameter for evaluation of clinical test performance. Validation is a laborious and time-consuming step in the NGS genetic testing. Fortunately, solutions exist to automate the process and implement in the laboratory workflow. Automated validation will not only save labs time and cost but also mitigate the risk of test failure.
Types of genetic test
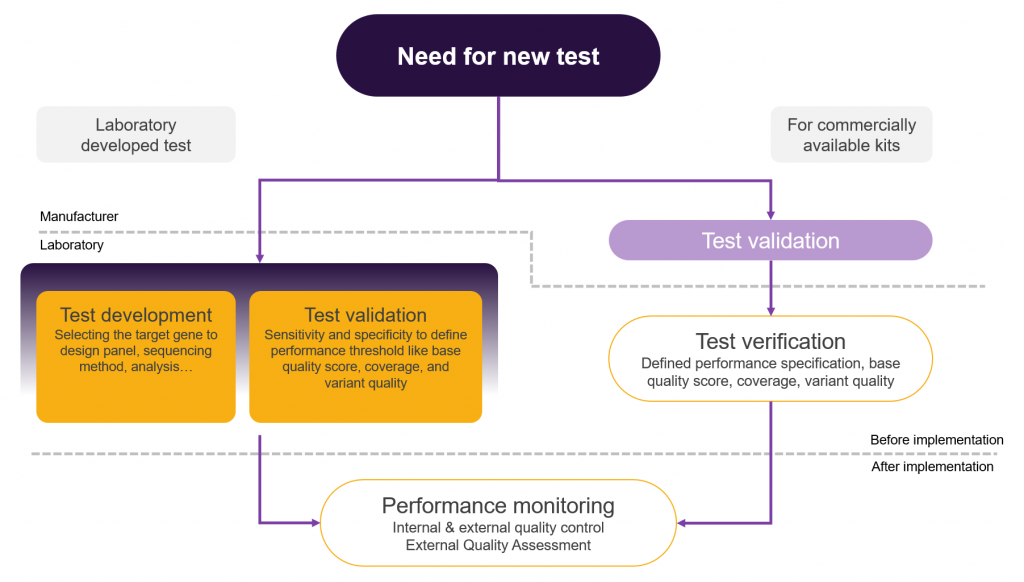
In this context, NGS-based genetic tests are of two distinct types: commercially available and laboratory developed tests (LDT). Both types need to be validated. For the LDTs, the responsibility for validation rests on the laboratory, while for commercially available tests it is the manufacturer who is responsible for the validation.
Table 1: Difference between LDTs and Commercially available test implementation
| Parameter | Laboratory developed tests (LDTs) | Commercially available tests |
| Regulation By | CLIA, ISO15189:2007 | Manufacturer: FDA for commercial approval Laboratory: CLIA, ISO15189:2007 for laboratory implementation |
| Requirement for Implementation | Validation, Performance monitoring | Manufacturer: Validation Laboratory: Verification, Performance monitoring |
| Time required to implement a new test in the lab | Potentially fast: In control of the laboratory | Potentially slow: Out of control of the laboratory |
| Cost focus for Lab implementation | On development and validation | On certified product price |
| Design | Free & customized | Fixed & limited choice |
| Multiplicity of tests | High | Low * |
Verification and Validation
Depending on the type of test labs perform, they have to either start from validation or from verification. The two are, however, not synonymous. Validation proves that a test has the capacity to perform as expected and that it can achieve the intended results. It additionally provides the performance characteristics of the measure(s) and the clinical efficacy of the test. On the other hand, verification is the process by which the laboratory verifies that it can replicate the established performance claims derived from the validation process. Thus, verification ensures the correct execution of the test before its implementation for diagnosis and can only be performed for validated tests.
Each test has to be validated and verified. However, commercially available tests, which have already been validated by the manufacturer still need to be verified by the lab before implementation as well as regularly after implementation, as required by quality assurance protocol.
Regulatory oversight for Validation and Verification
It should be noted that the term validation, although used widely by laboratories to mean establishing the performance characteristics for the test, may not appear in regulatory standards. The same holds true for verification which at times is termed as test evaluation (Table 2).
Table 2: Regulatory requirements for Validation & Verification

Framework for NGS test Validation
Validation of NGS tests for clinical implementation is challenging due to the complex nature of the technology. Fundamentally, NGS pipelines progress through three clearly distinguishable steps: 1. Preparation of the material for sequencing, 2. Sequencing, and 3. Bioinformatics analysis of the produced data. Validation needs to be carried out for each of these steps which makes it a time consuming process.
DNA extraction and library preparation
Due to the availability of different kits for nucleic acid extraction, fragment capture or amplification, validation of a NGS test ensures the molecules to be analysed are captured, and appear in the right proportion.
Sequencing
Different sequencing platforms apply different chemistries for nucleic acid detection and each detection method has its own biases and sequencing artefacts. Validation ensures the artefacts do not effect the test.
Bioinformatics
Bioinformatics workflows consist of wide variety of tools connected via their in- and outputs in a software environment. Workflows have to be optimised for different tasks due to the variability of sequence content for different genes. Validation ensures that the pipelines are able to detect the variants of importance in the genes of interest.
The implementation of NGS tests in a clinical laboratory is illustrated in Figure 1.
Parameters for NGS test validation
Fundamentally, genetic test validation is about the concordance of the observed variants with the base truth from a reference material or alternative methods like Sanger sequencing. Sensitivity and specificity are statistical measures of the performance of a binary classification function. These measures can be calculated from correctly identified variants (TP, true positives), correctly rejected (TN, true negatives), false alarms (FP, false positives), and missed variants (FN, false negatives). Other parameters can also be calculated (Table 3). Parameters to be calculated for validation depend on the type of test such as genotyping a particular mutation, finding out unknown mutations, type of mutations like SNP, INDEL, CNV, etc.. Validation is necessary in order to define optimal thresholds for the performance specification of a test.
Table 3 : Parameters for the NGS tests
| Validation Parameters | CAP3 | ACMG4 | EuroGentest | Nex-StoCT5 |
| Accuracy | X | X | X | |
| Precision | X | X | X | X |
| Analytical sensitivity / 1- False negative rate (ACMG) | X | X | X | X |
| Analytical specificity / 1- False positive rate (ACMG) | X | X | X | X |
| Reportable range & reference range | X | X | X | |
| Reproducibility and robustness | X | X | X | X |
Validation parameters for NGS tests:
Analytical sensitivity – Proportion of biological samples that have positive test results or known mutation and that are correctly classified as positive
Analytical specificity – Proportion of biological samples that have negative test results or no identified mutation (being tested for) and that are correctly classified as negative
Reportable range – The region of the genome in which sequence of an acceptable quality can be derived by the laboratory test
Reference range – Reportable sequence variations the assay can detect that are excepted to occur in an unaffected population
Accuracy – The degree of agreement between the nucleic acid sequenced derived from the assay and a reference sequence
Precision – The degree to which the repeated sequence analysis give the same result
Robustness – Likelihood of assay success
Reproducibility – the ability of an entire test to be duplicated with the same results
Performance monitoring
After validation and implementation of the test for the intended diagnostic purpose, verification needs to be done in order to monitor that the performance specification derived from the validation can be matched. The performance monitoring also includes internal and external quality control and is considered an ongoing verification of the test. The results of monitoring should be stored and analysed for further improvement of the test as well as development of new tests.
Conclusion
Due to various regulatory requirements, labs need to have proper validation and verification processes in place for NGS-based genetic tests. The introduction of a comprehensive and automated validation and monitoring process will not only help labs to get accreditation but also save time and costs in the daily monitoring procedures for the benefit of the overall quality and patient safety.
References:
- Matthijs G, Souche E, Alders M et al: Guidelines for diagnostic next generation sequencing. Eur J Hum Genet 2016; 24: 2–5.
- Molecular Pathology Checklist, CAP accreditation program 2014
- Aziz N, Zhao Q, Bry L, et al. College of American Pathologists’ Laboratory Standards for Next-Generation Sequencing Clinical Tests. Arch Pathol Lab Med 2014. doi:10.5858/arpa.2014-0250-CP
- Rehm HL, et al.; Working Group of the American College of Medical Genetics and Genomics Laboratory Quality Assurance Commitee. ACMG clinical laboratory standards for next-generation sequencing. Genet Med 2013;15:733–747.
- Gargis, A.S. et al. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat. Biotechnol 2012; 30, 1033–1036.
