Using Sample quality control in a laboratory introducing NGS testing
Validating first NGS tests and building the framework for regular NGS sample QC
The Challenge
Introducing NGS to a laboratory is no easy feat, requiring time, expertise and testing to ensure that the tests run by the laboratory are as trustworthy as possible, and providing data which can be relied upon when making diagnostic decisions.
Test validation is a key step in the process of getting a new test up and running and can prove particularly challenging in an environment where NGS is a new technology, and those working with the technology are in the process of transforming their genetics knowledge from other techniques to NGS.
For one laboratory, the challenge was a short timeframe for implementing routine NGS testing with an initial goal of being able to validate the first tests and introduce a consistent and repeatable quality control regime to enable routine clinical diagnoses with the data as soon as possible.
The Solution
Sample quality control from Euformatics was the ideal solution for the challenge. Combined with the specialist consultancy offered, the laboratory was able to validate its first test for the new NGS laboratory, while building its own knowledge of how to validate further tests in the future.
The validation process carried out by sample quality control focuses on ensuring a sufficient standard of sensitivity and precision of a test, based on assessing the coverage of a sample and performing a concordance analysis against a known truth-set.
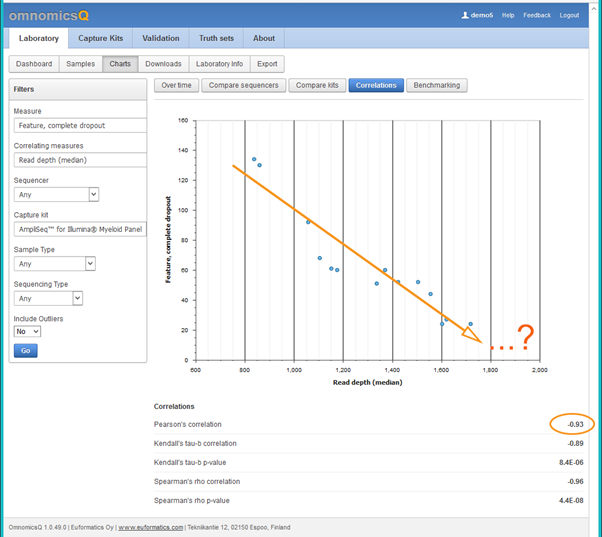
By being able to iteratively compare the read depth, the amount of DNA to load, as well as the coverage on target using automated coverage analysis combined with the charting functionality of sample quality control, it was possible to find the ideal ratio of DNA to read depth resulting in sufficient sensitivity and precision for the test. The results of the correlation between different tested read depths and coverage on target can be seen below in figure 1. The optimal read depth for this specific test was around 2000.
In addition, sample quality control was able to force an inspection of the BED file defining the region of interest for the kit being used, by highlighting a systematic error. The last 25 base pairs at the end of each amplicon were defined in the BED file, but never sequenced, and thus showed as uncovered regions at the end of each amplicon as they were absent from the sequence and alignment. Thanks to the automatic flagging of low coverage by sample quality control, the laboratory was able to spot the error, examine the alignments and rectify the fault in the BED file.
The Context
Validating an NGS test in a laboratory requires sufficient knowledge of the process and the right tools to ensure that validation can be carried out efficiently and with a high rate of reproducibility. For the majority of laboratories, it is also imperative to adhere to relevant ISO standards as they may also be looking to acquire official laboratory accreditation. In this case, one crucial aspect is the quality management documentation of the different processes that can affect the outcome for a patient. With sample quality control all the data required is readily and continuously at hand, both on a per sample basis and on a higher level, and triggers for automatic warning can be set.
For running a validation, the DNA amount loaded onto the sequencing chip is critical, since the sequencer manufacturer has routines for assessing the DNA amount based on observations on cluster size and signal intensity as well as other parameters obtained after the run. As a result, the DNA amount will also affect the coverage on target and can therefore also be monitored on the level of baits or amplicons and down to single base-pairs using sample quality control.
The read depth (DP) is another one of the most important parameters for obtaining sufficient measurement data to perform a proper variant calling. The required read depth will depend on multiple factors such as level of detection (LOD, the lowest variant allele frequency that must be detected), the complexity of the genomic ROI as well as the number of observations required to consider a called variant to be true.
By combining the automated functionality of sample quality control to routinely assess and measure the key parameters with its ability to both store the data for reporting purposes and chart it for comparisons, a laboratory is able to run an iterative process to arrive at the stage where their tests are running correctly and at a level where they can be sure that the data is trustworthy to be used in a diagnostics procedure.
About sample quality control
The sample quality control comprehensive NGS quality management system delivers the insights lab management and quality managers need to achieve top-tier confidence in sample data quality and lab performance by automating, centralising and contextualising quality control data management and analysis.
- Validate and verify assays with the assay validation module
- Apply widely to RNA and DNA (WGS to panels) with any kit, sequencer or platform.
- Track sample quality using traffic light notification for every sample according to stored user-defined SOPs.
- Report, chart and benchmark lab performance progress and changes.
- Compare performance to database of peers.
- Track performance of lab sequencers and kits.
- Implement CAP/AMP Standards and Guidelines, EuroGentest Guidelines and ISO 15189/17025 standards.