Key Highlights
We are now introducing omnomicsNGS version 2.13.0, bringing full support for somatic variant classification using the ClinGen/VICC/CGC guidelines, enhanced CIViC annotations, expanded structural variant analysis and flexible reporting workflows. This release delivers powerful new capabilities along with many refinements based on customer feedback and has a broader VCF compatibility for structural variants.
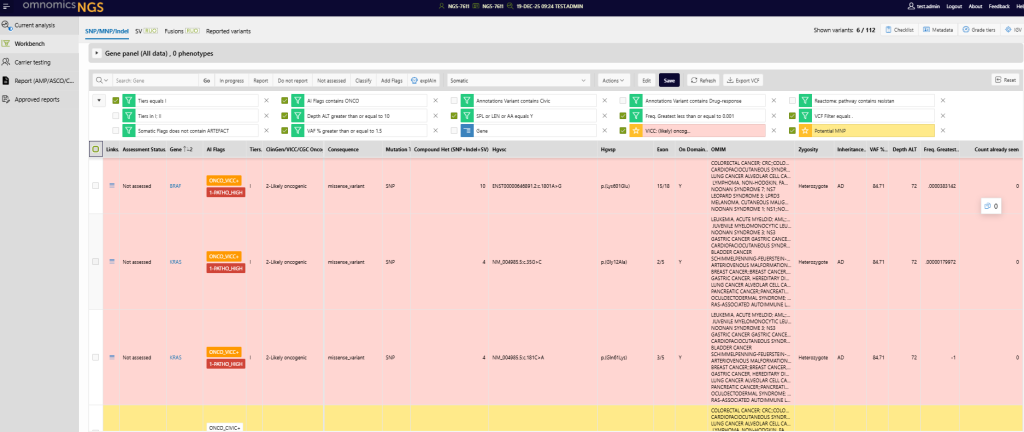
1 Improvements to somatic variant interpretation
1.1 New ClinGen/CGC/VICC oncogenicity classification
Gain deeper insights into somatic variants with automated oncogenicity classification based on the latest consensus guidelines. The system evaluates key evidence categories, applies standardised criteria, and provides transparent classification results with full editability and history tracking.
1.2 Expanded CIViC annotations including functional and oncogenic annotations
CIViC annotations now include functional impact and oncogenicity assessments, offering richer context for variant interpretation. Linking to each CIViC evidence makes it easier to review clinical relevance, supporting more confident decision-making.
2 Support for Structural Variants (SVs) and Uniparental disomy (UPD)
2.1 gnomAD 4.1 frequencies for CNVs
Copy-number variants (CNVs) now include population frequency data from gnomAD 4.1, enabling more accurate interpretation and easier distinction between common and rare events
2.2 Improved handling of INV, CTX, and breakend-based events
The platform now processes complex structural variants: Breakend-based SVs including inversions (INV), insertions (INS) and translocations (CTX) are properly grouped, annotated, and displayed
2.3 Enhanced LOH and compound heterozygosity analysis
Loss-of-heterozygosity (LOH) and compound heterozygous variants are detected and summarized on SNP/Indel and SV workbenches.
2.4 UPD detection with clearer region identification
Uniparental disomy events are now automatically detected. UPD can be visualised on the sample page with an editable option for setting up thresholds for the number of homozygous variants.
3 Broader VCF compatibility for structural variants
This version adds support for importing in particular structural variants (CNV, fusions, breakends) of various kinds, from VCF files of a wider range of variant callers, reducing duplication errors and ensuring accurate representation in the workbench for reliable analyses.
3.1 Combined SNP + InDel + CNV
Import combined SNP/Indel, CNV, and fusion variants seamlessly. This affects among other import from Thermo Fisher / Ion Torrent Genexus pipelines.
3.2 Fusions
Support for various fusion format interpretations of the VCF standard. This affects among other Arriba secondary RNAseq analysis, Dragen, Delly, and Oncomine Comprehensive Assay files.
3.3 Breakend SVs, tandem repeats, and other complex variants
Structural variants with breakends, inversions, translocations, or tandem repeats are now supported.
4 Automation & Command-Line Interface (CLI) support
We have further worked on automation of data input via the omnomicsNGS API. This allows automation of not only VCF file input, but also additional metadata. This facilitates computational processing of among other but not only additional non-vcf files with MSI, TMB, HRD information or sex and family structures for trio or larger analysis.
This affects the seamless ingestion of DRAGEN Somatic v4.3 and TSO500 output directories in a single step, but also allows a more automated transfer of any sample from a sequencer or a secondary pipeline to omnomicsNGS.
This feature has been a requirement in the EU-financed PCP Instand-NGS4P project on integration and standardisation of NGS Workflows for personalised therapy.
5 Improvements to Workbench
- AI flag-based filtering for faster and reliable detection of clinically relevant variant
- Real-time variant counts displayed before and after filtering
- Export filtered results directly to VCF for seamless downstream analysis
- Optimized user interface is now faster when changing workbench pages
- Structural variant display improvements
6 Improvements to Reporting
This version brings significant upgrades to ACMG/AMP and AMP/ASCO/CAP PDF reports:
- Updated styling, colors, and layout
- A separate SV/CNV table for clear presentation
- Addition of Quality Metric data (if available)
- EMA & FDA approved drug lists (for somatic report)
- Add ongoing and clinical trials with ability to filter based on country of recruitment and phase of the trial (for somatic report)
- Editable gene descriptions from NCBI (for somatic report)
- Language selection for PDF reports as an optional enhancement
7 Conclusion
Together, this release delivers faster, accurate, and reliable variant analysis and reporting, making it easier to turn data into actionable insights. If you have any questions, please contact us at support@euformatics.com.



