The newest version of omnomicsQ now contains support for detecting reads with technical or other contaminant sequences as a part of the raw (fastq) QC analysis. This functionality adds to the approximately 60 other QC metrics already available in omnomicsQ, and makes for a more comprehensive QC analysis of your NGS data. When creating a SOP, it is now possible to define what contamination database to use for the analysis, saving you time finding the contaminants separately, and integrating another key quality metric into your QC results.
What does this new feature look like?
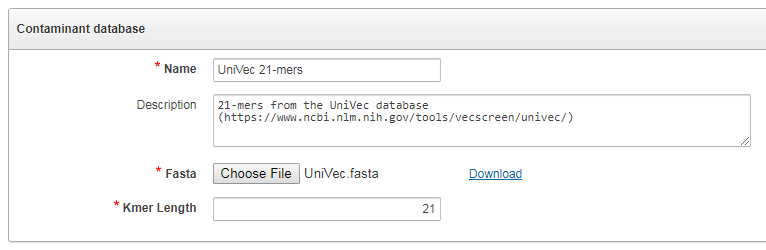
The contamination database is defined by a FASTA file containing sequences and the length of k-mers to be searched. Users can use e.g. the UniVec database, and specify for example k=21 as the k-mer length.

When running the fastq analysis, reads which contain at least one k-mer from the contamination databases are considered as “contaminated”, and the estimate of the percentage of contaminated reads is estimated.

To start using the contamination analysis feature, configure your SOPs with the contaminant k-mers you want to screen for and download the new version of the omnomicsQ desktop or command line client.
For more information on omnomicsQ, please get in touch at contact@euformatics.com or have a look at the omnomicsQ product page



