The omnomicsNGS variant interpretation, annotation, classification and reporting tool was upgraded to version 2.5 last week in a release that was packed with updates and new features that will be of particular benefit and interest to those doing WGS. We’ll take a look at the most important changes below and give you a quick overview of how they could help you in your work!
WGS filtering and analysis
Users of omnomicsNGS will already know that they have had the capability to handle WGS samples for a while but knowing where to start with a file that can contain millions of variants is often a challenge.
omnomicsNGS now features the ability to perform hard-filtering on samples before proceeding to analysis, allowing users to quickly filter out unwanted variants from the sample early in the process. Filtering can be done through setting a population frequency cut-off, uploading a BAM file containing a specified region, such as exomes, user-created gene panels or through phenotype associations using HPO terms.
The addition of this feature increases the speed that WGS samples can be analysed in the system, as it removes the need for frequent reannotations and allows users to only analyse the variants that are of most importance for the case at hand.

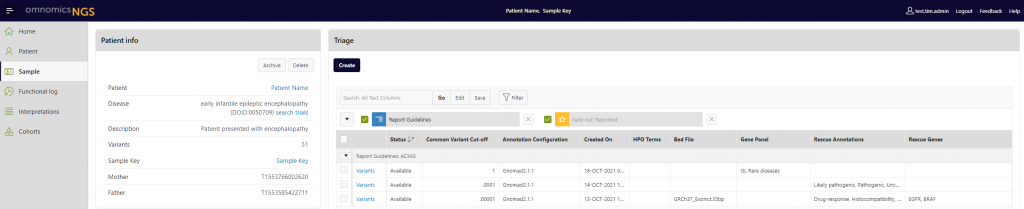
Multiple analyses per sample, saved in time
With the hard-filtering changes comes a new way of analysing samples in omnomicsNGS. Users can now create multiple analyses for the same sample and come back to them later or share with a colleague who also has access to the system. This gives you a much more flexible way of analysing samples in the software and offers a clearer way of navigating through the tool to get from VCF to clinical report.
It also means that the annotations associated with a specific analysis of a sample are stored as a snapshot in time and can be compared with later analyses using updated databases and annotations. For users who may need to re-analyse samples periodically as part of their quality management, this feature makes the comparison of samples and annotations possible.
Did you spot the Easter Egg?
The release also fixes several minor bugs, as well as an Easter Egg which some users may have noticed… In the list of integrated databases, Ensemble was listed instead of Ensembl. This has now been corrected, but if we have managed to miss any other small spelling errors, please do let us know! You can always drop us a message at contact@euformatics.com
See omnomicsNGS for yourself by booking your free demo and discover how the omnomics Suite can improve the processes in your laboratory



